Nuo Ivy Liu
Postdoctoral Research Fellow @ Goodarzi Lab, Arc Institute

email: ivliu@arcinstitute.org
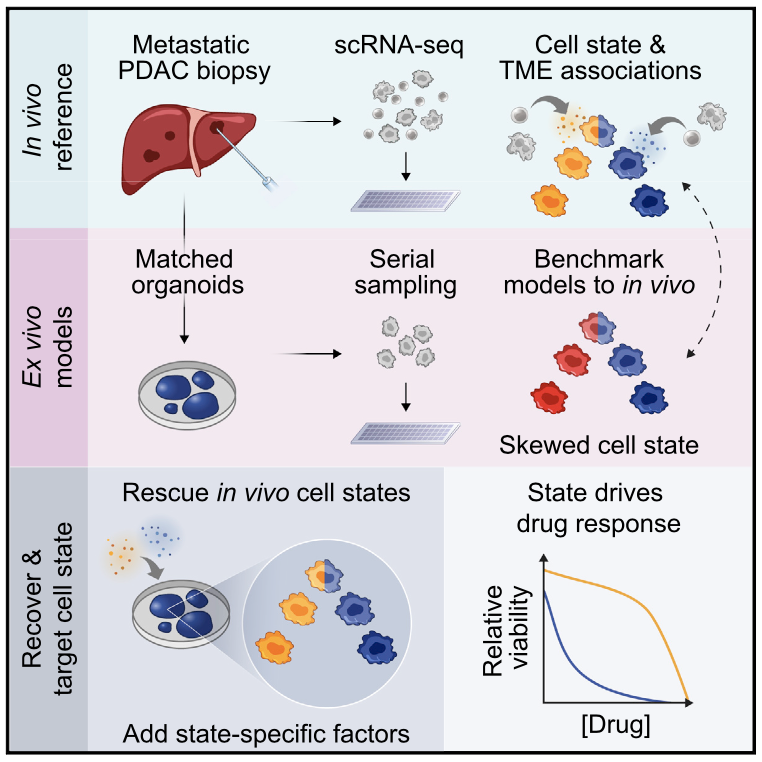
Hi! I am Nuo Liu (刘诺), people also know me as Ivy. I am a postdoctoral research fellow at the Goodarzi Lab at Arc Institute in Palo Alto, CA. My current research focuses on leveraging computational models to understand complex diseases and identify therapeutic targets, particularly vulnerabilities in cancers and Alzheimer’s disease. I also work on applying foundation models and LLMs to generate biomedical reasoning. I spend my time between New York and the Bay Area.
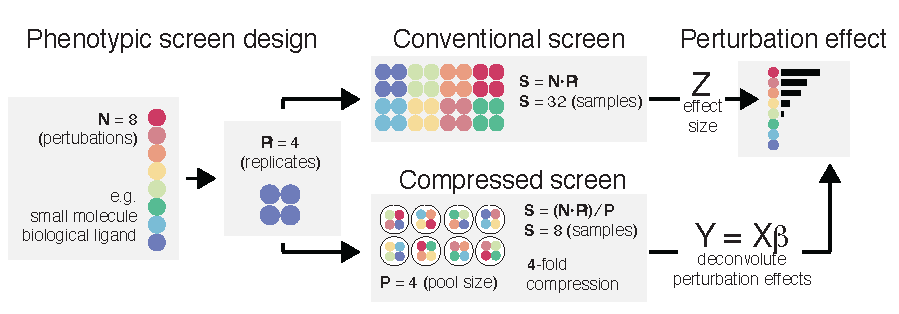
I completed my Ph.D. in Computational and Systems Biology (CSB) at MIT under the guidance of Professor Alex Shalek. My research at the Shalek Lab spanned multiple areas focused on the development and application of single-cell technologies (experimental and computational) to better understand perturbation responses in human cellular systems. I was supported by the NIH training grant and an internal graduate student fellowship through the Koch Institute at MIT. At MIT, I was involved with student organizations including MIT-CHIEF, Graduate Student Council External Affairs Board, and the graduate application assistance program for CSB.
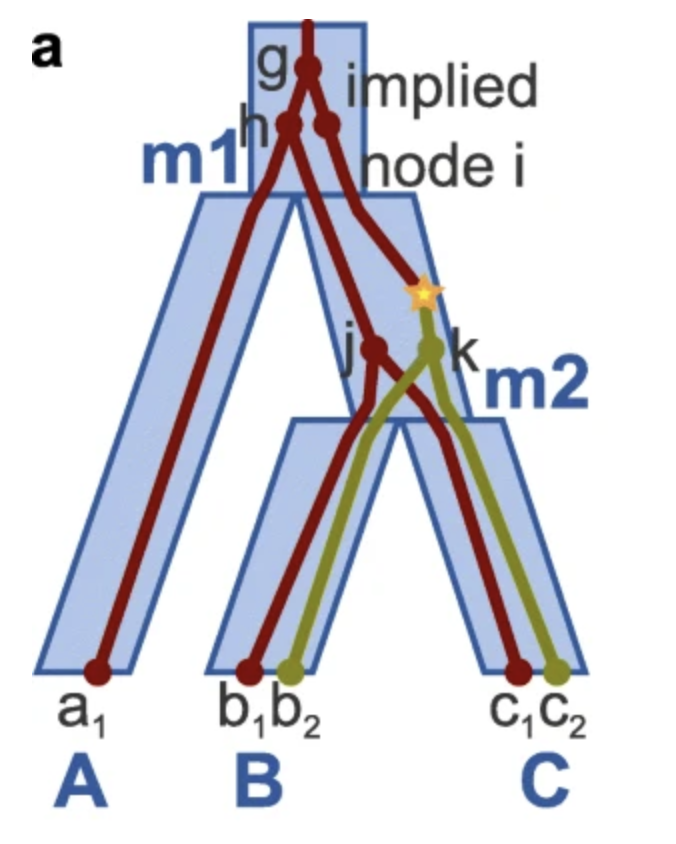
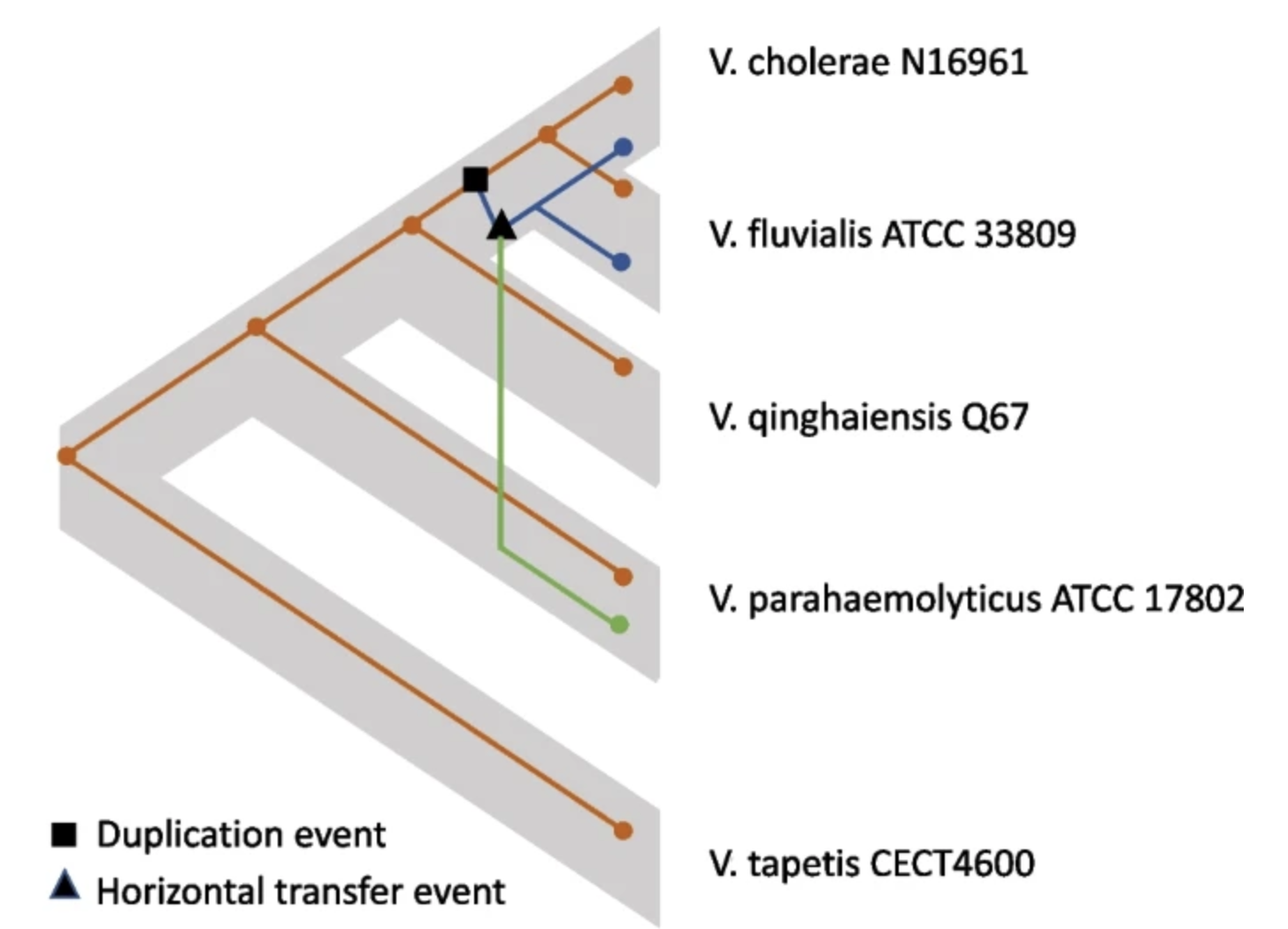
Before my Ph.D., I attended Harvey Mudd College, graduating with a B.S. in Mathematical and Computational Biology with honors in Computer Science and Biology. My interest in computational biology first started during college when I had the fortune to conduct research with Professor Catherine McFadden on evolutionary relationships of corals and with Professors Ran Libeskind-Hadas and Jessica Yi-Chieh Wu on algorithmic development for phylogenetic tree reconciliations. To gain more experience in translational research, I participated in summer research at Baylor College of Medicine and Texas Children’s Hospital through the SMART program, where I developed deep learning models for predicting epigenetic reprogramming events (supervised by Dr. Cristian Coarfa) and used linear programming to reconstruct transcriptional evolution of tumors (supervised by Dr. Pavel Sumazin). I was selected as a finalist for the Computing Research Association (CRA) undergraduate research award.
Outside of research, I enjoy pottery, painting, calligraphy, traveling, and hunting for the best matcha lattes.