Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer
Srivatsan Raghavan , Peter S. Winter , Andrew W. Navia , Hannah L. Williams , Alan DenAdel , Kristen E. Lowder , Jennyfer Galvez-Reyes , Radha L. Kalekar , Nolawit Mulugeta , Kevin S. Kapner , Manisha S. Raghavan , Ashir A. Borah , Nuo Liu, Sara A. Väyrynen , Andressa Dias Costa , Raymond W.S. Ng , Junning Wang , Emma K. Hill , Dorisanne Y. Ragon , Lauren K. Brais , Alex M. Jaeger , Liam F. Spurr , Yvonne Y. Li , Andrew D. Cherniack , Matthew A. Booker , Elizabeth F. Cohen , Michael Y. Tolstorukov , Isaac Wakiro , Asaf Rotem , Bruce E. Johnson , James M. McFarland , Ewa T. Sicinska , Tyler E. Jacks , Ryan J. Sullivan , Geoffrey I. Shapiro , Thomas E. Clancy , Kimberly Perez , Douglas A. Rubinson , Kimmie Ng , James M. Cleary , Lorin Crawford , Scott R. Manalis , Jonathan A. Nowak , Brian M. Wolpin , William C. Hahn , Andrew J. Aguirre , and Alex K. Shalek
Cell, Dec 2021
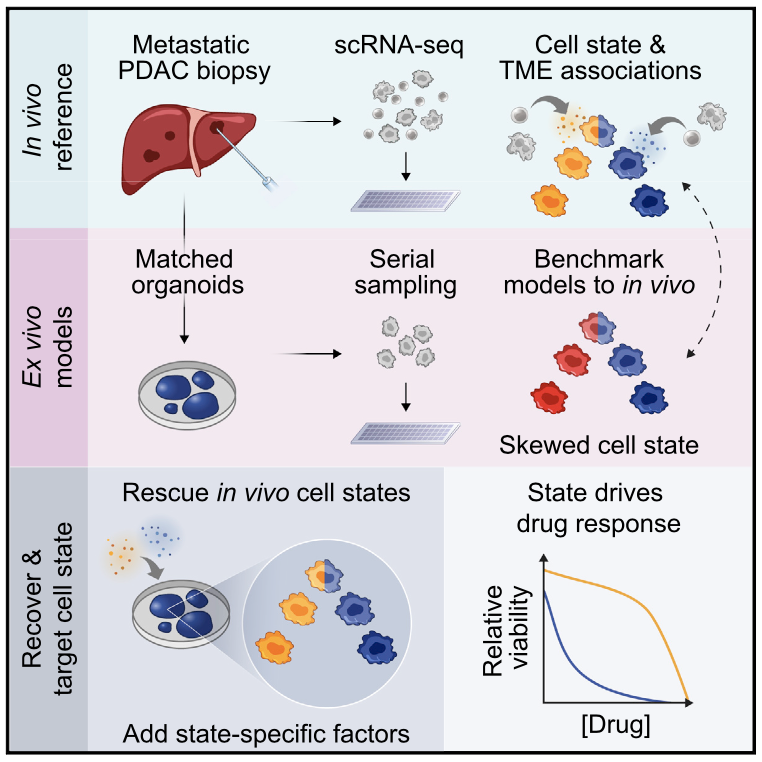
Summary Prognostically relevant RNA expression states exist in pancreatic ductal adenocarcinoma (PDAC), but our understanding of their drivers, stability, and relationship to therapeutic response is limited. To examine these attributes systematically, we profiled metastatic biopsies and matched organoid models at single-cell resolution. In vivo, we identify a new intermediate PDAC transcriptional cell state and uncover distinct site- and state-specific tumor microenvironments (TMEs). Benchmarking models against this reference map, we reveal strong culture-specific biases in cancer cell transcriptional state representation driven by altered TME signals. We restore expression state heterogeneity by adding back in vivo-relevant factors and show plasticity in culture models. Further, we prove that non-genetic modulation of cell state can strongly influence drug responses, uncovering state-specific vulnerabilities. This work provides a broadly applicable framework for aligning cell states across in vivo and ex vivo settings, identifying drivers of transcriptional plasticity and manipulating cell state to target associated vulnerabilities.